The pathway design phase, which represents the backbone of our project, has been completed during the first year. We have now almost completed the first two phases of the project (in silico and in vitro) and we are moving towards the 3rd phase in vivo. We have first implemented the pathways in the bacteria E. coli using it as a platform for the pathway selection. This step is paving the way for the introduction of the most promising pathways into the final organisms: cyanobacteria and plants. Below you can see two of the selected pathway:
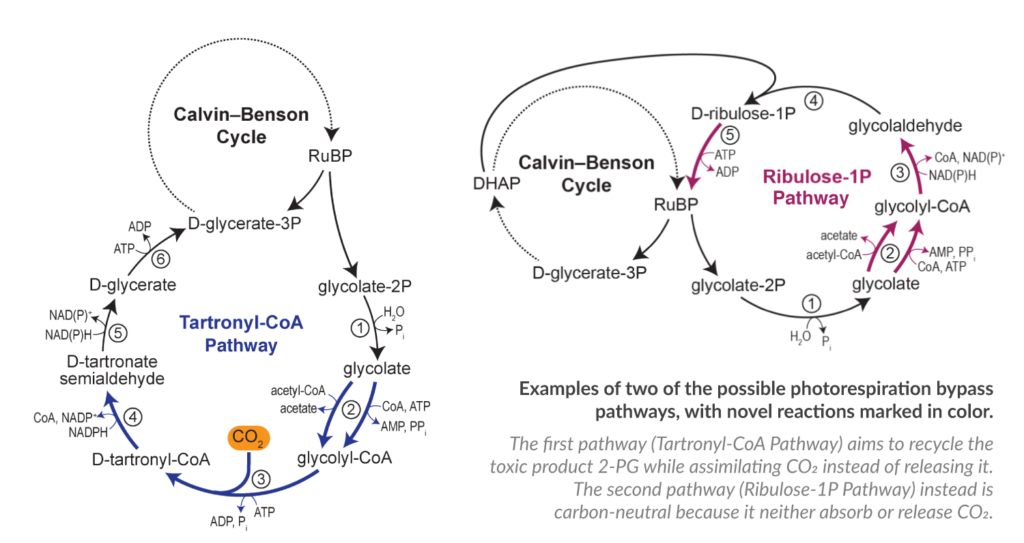
During the past year, the FutureAgriculture team have worked strongly on the carbon positive pathway which has the potential to increase plant yield by 20% according to the predictions. We have identified and analysed in detail all the building blocks necessary to the pathway, which means the enzymes that perform the reactions.
The final selection of enzymes has been already tested within the in vitro plant metabolism and the outcomes have been very encouraging. We have the first proof of principle that at least in vitro the pathway directly improves CO2 fixation metabolism of the plant and that the pathway if transplanted in vivo, has the potential to positively affect plant growth.
The selected enzymes have been already passed to the partners at Evogene and Imperial College London to be tested in plants and cyanobacteria. Currently, the consortium is busy refining all the details to make sure that the enzymes can perform their function in vivo too. We are all eager to see the results in vivo because there can always be surprises – that’s why it’s important to go back and forth between models and the experiments and we will refine the final pathway according to the results in vivo.
Our results in a nutshell:
 We identified more than 100 candidate pathways that can potentially bypass the natural photorespiration without releasing CO2. The pathways contain new plausible reactions catalyzed by modified enzymes.
We identified more than 100 candidate pathways that can potentially bypass the natural photorespiration without releasing CO2. The pathways contain new plausible reactions catalyzed by modified enzymes.
 We have engineered existing promiscuous enzymes that efficiently catalyze the novel reactions required by four of the selected synthetic pathways. They are currently undergoing the evolution stage.
We have engineered existing promiscuous enzymes that efficiently catalyze the novel reactions required by four of the selected synthetic pathways. They are currently undergoing the evolution stage.
 Our ability to test pathways in vitro and in vivo is limited. We have developed a computational model of plant metabolism to filter them out taking into account the main reactions involved in photosynthesis.
Our ability to test pathways in vitro and in vivo is limited. We have developed a computational model of plant metabolism to filter them out taking into account the main reactions involved in photosynthesis.
 It’s not enough to find an enzyme that can perform a reaction; the enzyme needs to be sufficiently active too. At FA we have worked to improve all the selected enzymes.
It’s not enough to find an enzyme that can perform a reaction; the enzyme needs to be sufficiently active too. At FA we have worked to improve all the selected enzymes.
 Once identified the best enzymes, it’s time to study them in their context: the carbon positive CO2 fixation pathway. It’s important that the enzyme can work as a team.
Once identified the best enzymes, it’s time to study them in their context: the carbon positive CO2 fixation pathway. It’s important that the enzyme can work as a team.
 So far, to improve enzyme efficiency the active site has been taken into account. FA team is using high-throughput mutagenesis to expand the modifications to the whole structure.
So far, to improve enzyme efficiency the active site has been taken into account. FA team is using high-throughput mutagenesis to expand the modifications to the whole structure.
 Who would have known that an unnoticeable organism as a bacterium would be the father of photosynthesis? Cyanobacteria are an ideal model organism to study photorespiration.
Who would have known that an unnoticeable organism as a bacterium would be the father of photosynthesis? Cyanobacteria are an ideal model organism to study photorespiration.
 At the ICL, scientists use cyanobacteria to evaluate the in vivo behaviour of the synthetic pathways. They carefully evaluate their impact on carbon fluxes, metabolism and cell growth.
At the ICL, scientists use cyanobacteria to evaluate the in vivo behaviour of the synthetic pathways. They carefully evaluate their impact on carbon fluxes, metabolism and cell growth.
 Introducing new genes into cyanobacteria is quicker than into plants but not as quick as one might expect. Read on to discover the challenges of introducing synthetic pathways.
Introducing new genes into cyanobacteria is quicker than into plants but not as quick as one might expect. Read on to discover the challenges of introducing synthetic pathways.
 Evogene works with two different model plants in FutureAgriculture; together represent two major types of crop plants
Evogene works with two different model plants in FutureAgriculture; together represent two major types of crop plants
 To implement the pathways in higher plants, Evogene takes advantage of the pathogenic machinery of Agrobacterium tumefaciens.
To implement the pathways in higher plants, Evogene takes advantage of the pathogenic machinery of Agrobacterium tumefaciens.
 Evogene is growing and testing the plants in its greenhouses with the final aim of validating their growth-advantage.
Evogene is growing and testing the plants in its greenhouses with the final aim of validating their growth-advantage.
 We have identified two groups of promising synthetic photorespiration bypasses. The most promising pathways have been tested in vitro and in vivo.
We have identified two groups of promising synthetic photorespiration bypasses. The most promising pathways have been tested in vitro and in vivo.
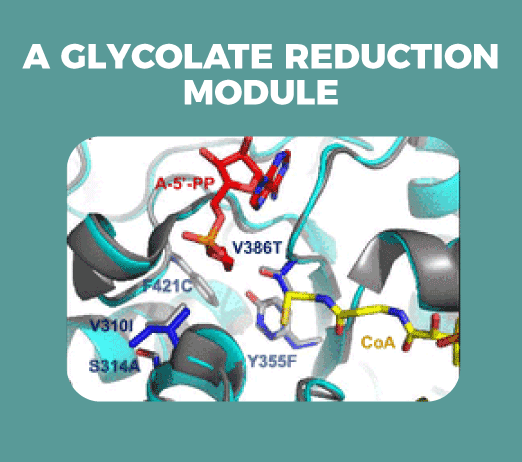 By means of computational design and directed evolution, we have engineered a common module to several pathways: the reduction of glycolate to glycolaldehyde.
By means of computational design and directed evolution, we have engineered a common module to several pathways: the reduction of glycolate to glycolaldehyde.
 The MPI-MP team has developed metabolic sensor strains which link the bacterial growth to the presence of a compound, replacing screening with direct selection.
The MPI-MP team has developed metabolic sensor strains which link the bacterial growth to the presence of a compound, replacing screening with direct selection.






