We integrate diverse skills in computational biology, chemistry, biochemistry, microbiology, and plant physiology, for the design and analysis of novel routes, the engineering of novel enzymatic reactions, and the implementations of optimized, synthetic metabolic pathways in plants.
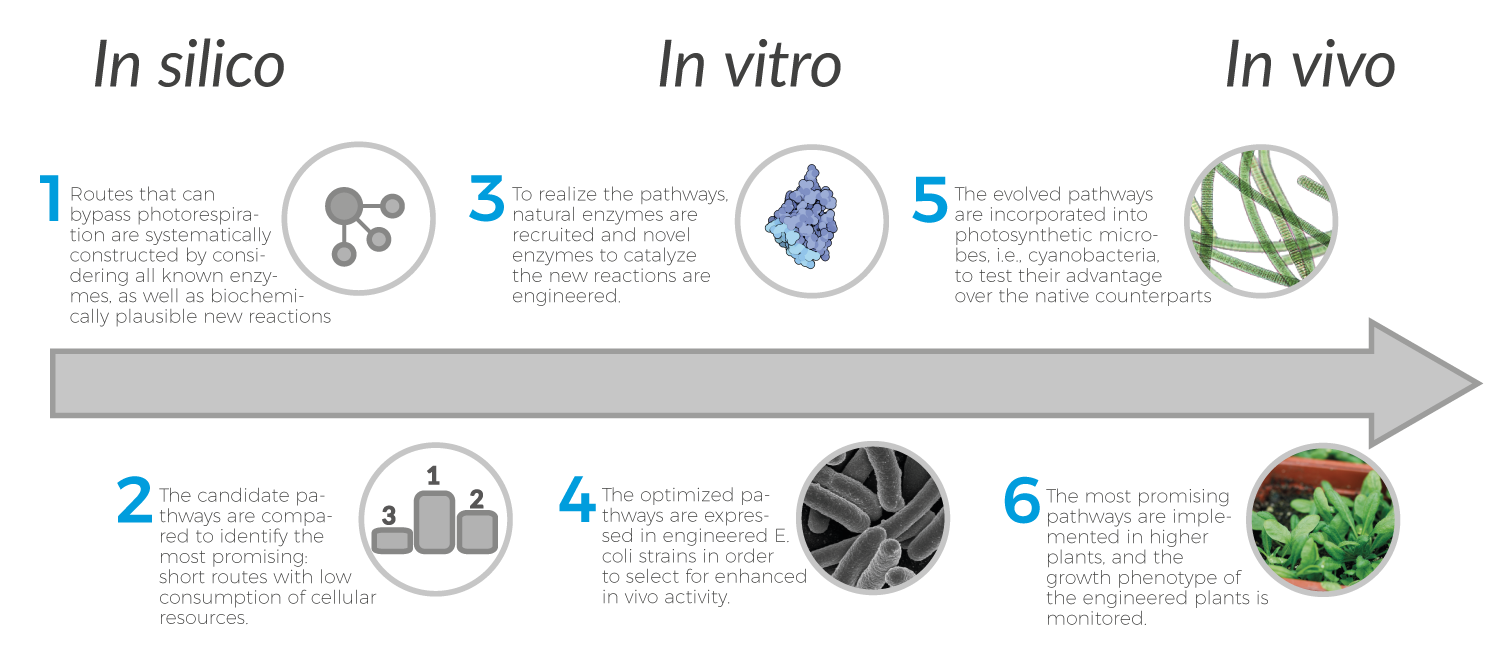
While the improvement of photosynthetic rate and yield via transgenic approaches has been a hot research topic for many years, these efforts focused on introducing existing pathways into new plant hosts. We adopt a radically different approach. Rather than reshuffling and grafting existing components in a fashion that resembles natural evolution and is in line with current metabolic engineering thinking, we systematically explore novel routes that cannot be obtained by mixing-and-matching of existing, natural enzyme components. Our approach demands the de novo engineering of new enzymes to catalyze metabolic transformations that are unknown in nature. These synthetic enzymes are integrated with existing ones to obtain de novo pathways optimized by chemical logic, which will be in turn realized within bacteria and plants.
Importantly, given the combinatorial nature of metabolic pathways, the addition of only one novel reaction dramatically expands the solution space of possible pathways, thus fully realizing the potential of synthetic biology. Yet, so far, only a handful of studies implemented synthetic pathways that harbor novel reactions. Here, we take this strategy to a new level, by constructing de novo pathways within the very core of carbon metabolism: plant photorespiration.






